Alignment of Clinical Trial Data for Pharmaceuticals
Industry: Life Sciences, Pharma & Biotech
Focus Area: Alignment of Clinical Trial Data
Background
A global pharmaceutical company received datasets with the results of drug trials from different partners and sites.
Each dataset described medical conditions, diagnosis and outcomes using terminologies local to the party or system that created it, making it difficult to reliably aggregate and analyze results.
Challenge
Aggregating and analyzing data requires terminology alignment. This is a challenging task because:
- Even when all data comes from the same source, terminology used may be inconsistent.
- Modern day clinical trial data is becoming increasingly more complex and heterogeneous.
- All data is no longer collected from a single system or via unified case report forms (CRF).
Multiple sources, and/or streams of data, are now commonplace as the science of clinical investigation is becoming more sophisticated. For example, data sources may include: wearable devices, specialty lab data, imaging data, -omics (genomics, etc.) data, companion diagnostics, healthcare data, real world evidence, and disease and patient registries. Each of these sources and/or streams may be using different terminologies.
The company needed to align, normalize and cleanse received information prior to importing it into downstream systems for analysis and reporting.
Solution
The company decided to use TopBraid EDG to cleanse and curate the datasets.
First, each clinical drug trial study was described in EDG as a subject area. Owner and a curating team were assigned. With the study established, the datasets for a study are being imported as they come in from the field. Data is imported from CSV files and other structured formats.
TopBraid EDG was used to create for each study, from data in the datasets, study-specific glossaries of terms for diseases, tissues and other relevant concepts.
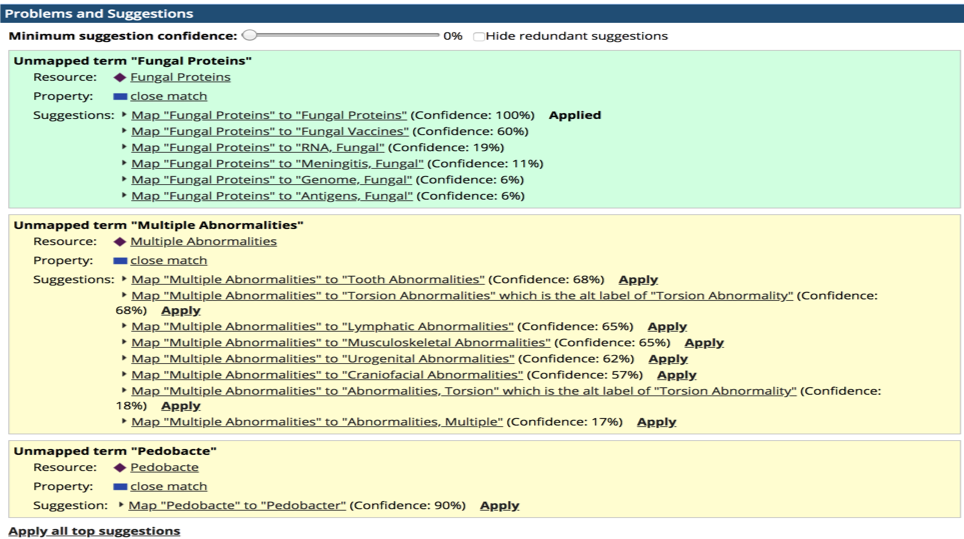
It then automatically analyzed these “local terminologies” and found suitable matching terms from the industry standard vocabularies such as SNOMED, MESH, Entrez Gene, NCIt as well as the client’s own standard controlled vocabularies of relevant terms.
Doing this efficiently, correctly and seamlessly ensures reliable analysis of clinical trial data while keeping clinical development costs in check, and clinical development timelines nimble.
When finding suitable terms, TopBraid EDG assigns a probability to each match found (Figure 1).
Ingest, auto-match and enrich processes can be invoked by users on demand or executed as scheduled services.
Users could then request TopBraid EDG to enrich the original datasets by adding or replacing data as deemed necessary. The normalized datasets are available for retrieval by users or for provisioning to downstream systems.
Results
After the terms in the glossaries were auto-matched to the standard vocabularies and validated, results of the matching processes were saved as crosswalks for further re-use.
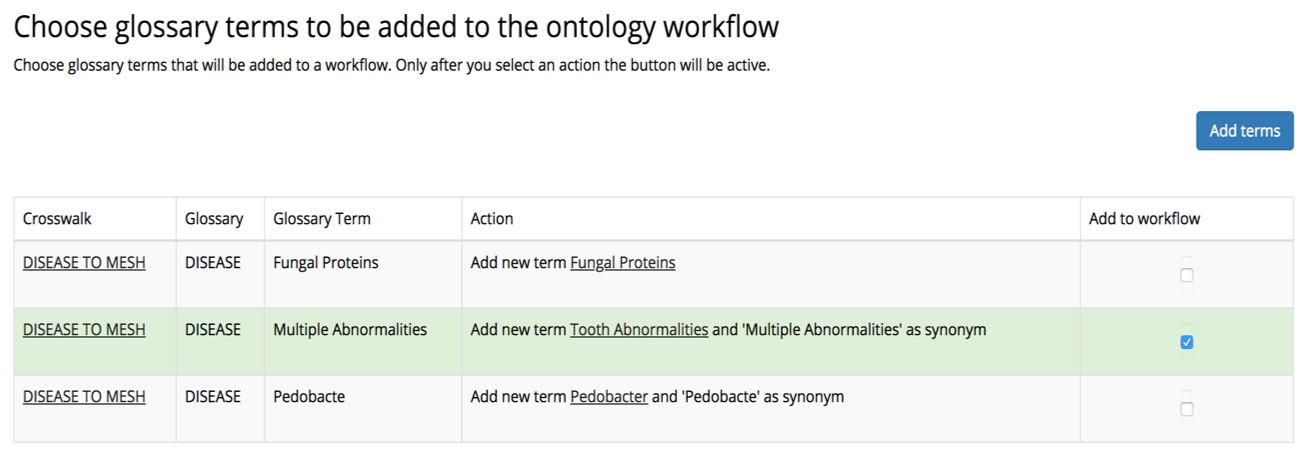
Figure 2 illustrates how data curators can decide to extend the client’s standard vocabulary of relevant terms by adding to it terms that were found in a study-specific glossary and matched to the industry standard, but do not yet exist in the company’s standard.


